 |
| Home | Research | Publications | Prospective Students | Teaching | Contact | Physics | Resources | Fun |
| Research Home
Current Projects Other Projects |
Welcome to the Plotkin Research Group!

Plotkin Group members, Left to right: Pranav Garg, Kathryn Lande, Aina Adekunle, Kyra Boulding, Dr. Steven Plotkin, Dr. Luke McAlary, Shawn Hsueh, Lana Kashino, Tiam Heydari ( Not shown: Mine Sher)
Come meet the members of our group, review our current research, and find out how you can join, We are currently looking for experimental researchers in either the developmental biology or protein misfolding projects below.
Novel computational approaches to predict misfolding-specific epitopes and design precision immunotherapeutics for neurodegenerative disease
We have developed a new way to rationally-predict disease-specific epitopes in misfolding prone proteins. I am co-inventor on >50 patents in the last 6 years on applications of this technology to personalized medicine in Alzheimer's disease (AD), Parkinson's disease (PD), and amyotrophic lateral sclerosis (ALS) immunotherapy, along with my experimental colleague Neil Cashman. The
inventions focus on computationallypredicted diseasespecific epitopes in Amyloid beta and tau protein, two of the primary proteins whose misfolding and aggregation is implicated in AD. We have designed and developed conformationallyselective antibodies (Abs) that selectively bind these conformational epitopes. These precision Abs are designed to spare healthy protein, but target misfolded protein when it is behaving pathogenically. They are selective to toxic oligomers. We also pursue optimal epitopes and precision Abs for other proteins and diseases, including
alphasynuclein in PD, and TDP43 in ALS.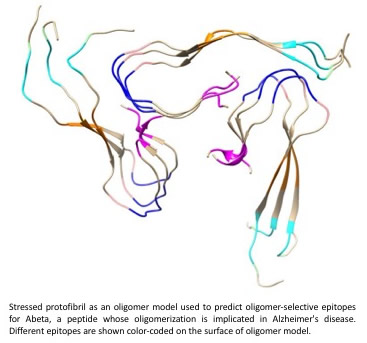
The methodology involves stressing structured fibrils computationally using a “Collective Coordinates” method (see this publication: PDF and also "Systems and Methods for Predicting Misfolded Protein Epitopes by Collective Coordinate Biasing" S.S. Plotkin, PCT/CA2016/051306).
Variations on irrational design are often used as a best guess for antibody development. I emphasize that rational design of antibodies from first principles using concepts from physical chemistry and structural molecular biology is a transformative concept in neurodegenerative disease immunotherapy, whose consequences are only beginning to be explored, and are being pioneered by our lab.
Given epitope predictions, we develop precision antibodies that best target diseased protein. Epitopes are optimally presented in cyclic peptides again using a computational design protocol which we have pioneered. Our Abs will ignore an epitope if the conformation is the same as in healthy protein, and our Abs are also designed to spare off-pathway targets in the human proteome. We use a multiplecriteria decision making (MDCM) scheme to screen and discover optimal precision Abs.
This work has been done in close collaboration with my colleague Neil Cashman, in experimental neurology. The IP has been supported and licensed by ProMIS Neurosciences Inc., a development-stage biotech company developing precision therapeutics for the treatment of AD and ALS. I am the Chief Physics Officer for ProMIS Neurosciences (and maybe the only Chief Physics Officer anywhere!)
A novel feature of our AD antibodies directed against A-beta, which has resulted from our unique computational design approach, is that they do not bind Abeta plaques. This has been verified by immunohistochemistry of normal and AD patient brain sections. This is important, because most healthy but aged individuals have brains that are abundant with plaque, but they are free from AD symptoms because they do not carry toxic oligomeric strains of Abeta. The oligomer-selectivity of our antibodies is a unique strength: Many current commercial antibody hopefuls do in fact bind plaque, and as a result, treatment with these Abs may suffer from doselimitations related to the inflammation and edema they induce.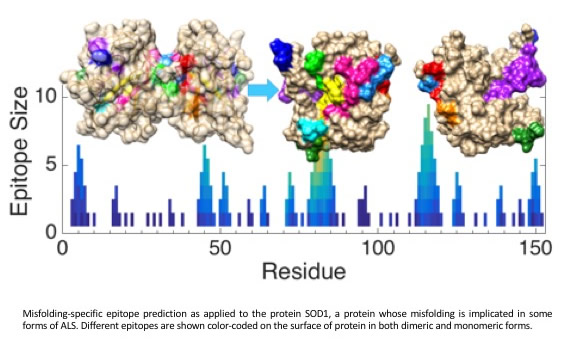
Designing Abs that are conformationally-selective to toxic oligomers is an extremely
powerful technological approach, which has the potential to transform the way
therapies are developed in the pharmaceutical industry, with profound implications
for human health benefits of both Canadians and people abroad.
For related publications see here.
Molecular-Genetic Origins of Multicellularity 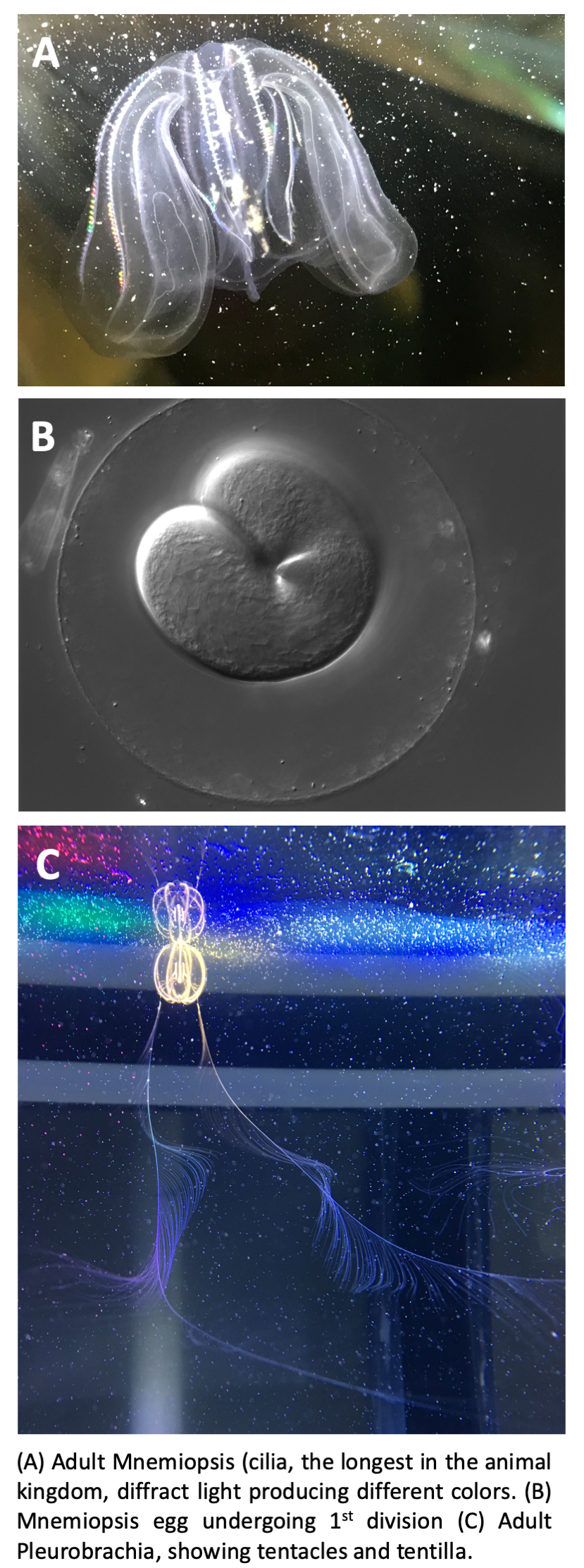
The embryologic question of how a multicellular organism can arise from a single cell is intimately linked with the equally fundamental evolutionary question of how complex multicellular organisms arose from single-celled ancestors. The origin of multicellularity is a pivotal transition to complex life, as it was a necessary precursor to higher life forms containing complex organs including brains. However, it remains enigmatic as most of the molecular, genetic, and evolutionary mechanisms involved in the transition are only beginning to be unraveled.
Multicellularity emerges when cells cooperatively differentiate and organize spatially into an integrated organism in a process that is genetically encoded for successive generations to reliably reproduce from a single progenitor cell. Depending on the degree to which cellular aggregation, sustained cell-to-cell inter-connection, communication, and cooperation are integrated, the transition to multicellularity has occurred anywhere from about a dozen to about 40 independent times across the tree of life during evolutionary history. The repeated instances of this transition across distinct environments and various epochs, and on different phylogenetic backgrounds, suggests that multicellularity is a phenomenon that arose, and can ?arise, as a generic physicochemical response to various environmental pressures. In this sense then it is a natural consequence of evolution, and a universal aspect of life.
This is a new project that has become one of the primary focuses of our lab. We will investigate the function and evolution of genetic regulatory networks (GRNs) involved in the process of cell differentiation; we are currently using CRISPR/Cas9 genetic manipulation methods in model and non-model organisms to address this question.
One extant animal lineage that has emerged as a candidate for the sister group to the other metazoa are the Ctenophora, which diverged from other animal clades over 550 million years ago: Ctenophores or comb jellies are a phylum of gelatinous zooplankton found in all of the world’s oceans. We focus on a representative ctenophore to obtain information on the conservation of relevant gene regulatory networks (GRNs) across the metazoans, to address questions of the GRN’s evolutionary origins. The ctenophore Mnemiopsis leidyi is in many ways an ideal embryologic system to investigate questions on the origins of animal multicellularity.
Home | Research | Publications | Prospective Students | Teaching | Contact | Physics | Resources || Fun |